
-
中文 | EN

中文 | EN
学历:博士研究生
职称:研究员
邮箱:yzhang@mail.sioc.ac.cn
电话:021- 68582386
地址:上海市海科路100号13号楼432
2006年9月-2010年6月 四川大学生物学基地班 学士学位
2010年9月-2015年7月 清华大学生命学院 博士学位
2015年10月-2020年10月 美国洛克菲勒大学 博士后
2020年10月-2021年3月 美国洛克菲勒大学 助理研究员
2021年3月-至今 中国科学院生物与化学交叉研究中心 研究员、课题组长
Professor Yixiao Zhang’s long-term passion is to use single-particle cryo-EM to answer challenging mechanistic questions. Biological macromolecules are of fundamental importance for cell physiology. Studies aimed at elucidating the mechanisms underlying the biological functions of these biomacromolecules are thus key to our understanding of basic biological processes and how their dysfunction results in disease. The goal of structural biology is to visualize the architecture of biomacromolecules and to establish the molecular basis that enables them to perform their functions.
While there has been tremendous progress in the use of single-particle Cryo-EM for structure determination, limitations and challenges remain. For example, it can be difficult to recapitulate the appropriate physiological environment for a protein and it can be exquisitely difficult to resolve the conformation of an underrepresented class of molecules in a structurally heterogeneous population. These issues impede the structural study of many important biomacromolecules. Professor Zhang has developed novel methods to mimic membrane tension to study mechanosensitive ion channels, and overcame numerous challenges on diverse biological samples, ranging from membrane proteins, protein-protein complexes, protein-RNA complexes to intact virus. These investigations elucidated several exciting and long-awaited functional mechanisms and resulted in several papers in top-tier journals, including Nature (2021), Science (2021), Molecular Cell (2020a, 2020b), Nature Structural & Molecular Biology (2014), Nature Communications (2021), and PNAS (2017a,2017b).
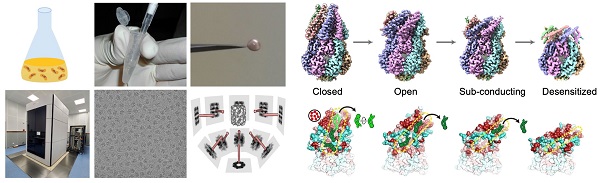
Currently Professor Zhang’s group is working on:
(1) mammalian mechanosensitive channels to understand the human sensations and other biological processes stimulated by mechanical force.
(2) the machineries that involved in neurodegeneration and other diseases to understand their molecular mechanisms.
(3) the biological processes that govern X-chromosome inactivation.
本课题组致力于运用冷冻电镜技术研究富有挑战性的生物学机制问题。蛋白质、核酸、脂类等生物大分子是体内各种生命活动的主要参与者,他们在体内精密协同,执行各种不同的生理功能。结构生物学的研究通过解析这些生物大分子精细的三维结构,阐释它们发挥功能的分子机制,并为相关药物的研发和疾病的治疗提供基础。冷冻电镜技术在近十年来发展十分迅速,已成为生物大分子结构解析的主要手段。本课题组将综合利用冷冻电镜技术、生物化学、细胞生物学、神经生物学等手段研究重要生物大分子的结构与功能。
主要研究方向包括:
(1)与触觉、听觉等感知觉形成相关的机械力通道机制研究
(2)与神经退行疾病相关的生物大分子复合物结构与功能研究
(3)X染色体失活机制研究
1. Han, Y., Zhou, Z., Jin, R., Dai, F., Ge, Y., Ju, X., Ma, X., He, S., Yuan, L., Wang, Y., Yang, W., Yue, X., Chen, Z., Sun, Y., Corry, B.# , Cox, C. D.#, and Zhang, Y.# (2024). Mechanical activation opens a lipid-lined pore in OSCA ion channels. Nature, 1-9.
2. Zhou, Z., Ma, X., Lin, Y., Cheng, D., Bavi, N., Secker, G.A., Li, J.V., Janbandhu, V., Sutton, D.L., Scott, H.S., Zhang, Y. #, Cox, C.D.# (2023). MyoD-family inhibitor proteins act as auxiliary subunits of Piezo channels. Science, 381, 799-804.
3. Yuan, L., Han, Y., Zhao, J., Zhang, Y.#, and Sun, Y#. (2023). Recognition and cleavage mechanism of intron-containing pre-tRNA by human TSEN endonuclease complex. Nature Communications, 14, 6071.
4. Xia, A., Wang, X., He, J., Wu, W., Jiang, W., Xue, S., Zhang, Q., Gao, Y., Han, Y., and Li, Y., Peng, X., Xie, M., Mayer, C. T., Liu, J., Hua, C., Sha, Y., Xu, W., Huang, J., Ying, T., Jiang, S., Xie, Y., Cai, Q., Lu, L. #, Silva, I. T. #, Yuan, Z. #, Zhang, Y. #, and Wang, Q#. (2023). Cross-reactive antibody response to Monkeypox virus surface proteins in a small proportion of individuals with and without Chinese smallpox vaccination history. BMC biology, 21, 205.
5. Zhang, Y., Daday, C., Gu, R., Cox, C.D., Martinac, B., Groot, B., Walz, T. (2021). Visualizing the mechanosensitive ion channel MscS under membrane tension. Nature, 590, 509-514.
6. Dan, J.*, Zhang, Y.*, Lee, C.H., Valencia-Sanchez, M., Zhang, J., Wang, M., Holder, M., Svetlov, V., Tan, D., Nudler, E., Reinberg, D., Walz, T., Armache, K. (2021). Structures of monomeric and dimeric PRC:EZH1 reveal flexible modules involved in chromatin compaction. Nature Communications, 12(1): 714.
7. Sun, Y.*, Zhang, Y.*, Aik, W.S., Yang, X.-C., Marzluff, W.F., Walz, T., Dominski, Z., and Tong, L. (2020). Structure of an active human histone pre-mRNA 3’-end processing machinery. Science 367, 700-703.
8. Zhang, Y.*, Sun, Y.*, Shi, Y., Walz, T., and Tong, L. (2020). Structural Insights into the Human Pre-mRNA 3’-End Processing Machinery. Molecular Cell 77: 800-809.
9. Su, M.*, Zhu, L.*, Zhang, Y.*, Paknejad, N.*, Dey, R., Huang, J., Lee, M.Y., Williams, D., Jordan, K.D., Eng, E.T., Ernst, O.P., Meyerson, J., Hite, R., Walz, T., Liu, W., Huang, X. (2020). Structural basis of the activation of heterotrimeric Gs-protein by isoproterenol-bound b1-adrenergic receptor. Molecular Cell 80:59-71.
10. Nešić, D.*, Zhang, Y.*, Spasic, A.*, Li, J.*, Provasi, D., Filizola, M., Walz, T., and Coller, B.S. (2020). Cryo-Electron Microscopy Structure of the αIIbβ3-Abciximab Complex. Arteriosclerosis, Thrombosis, and Vascular Biology. 119.313671.
11. Yu, J.*, Zhang, B.*, Zhang, Y.*, Xu, C.Q., Zhuo, W., Ge, J., Li, J., Gao, N., Li, Y., and Yang, M. (2018). A binding-block ion selective mechanism revealed by a Na/K selective channel. Protein & Cell 9, 629-639.
12. Lees, J.A.*, Zhang, Y.*, Oh, M.S., Schauder, C.M., Yu, X., Baskin, J.M., Dobbs, K., Notarangelo, L.D., De Camilli, P., Walz, T., Renisch, K. (2017). Architecture of the human PI4KIIIalpha lipid kinase complex. Proc Natl Acad Sci. U.S.A 114, 13720-13725.
13. Sun, Y.*, Zhang, Y.*, Hamilton, K., Manley, J.L., Shi, Y., Walz, T., and Tong, L. (2017). Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc Natl Acad Sci. U.S.A 115, E1419-E1428.
14. Zhang, Y., Ma, C., Yuan, Y., Zhu, J., Li, N., Chen, C., Wu, S., Yu, L., Lei, J., and Gao, N. (2014). Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Nature Structural & Molecular Biology 21, 1042-1046.
15. Wu, S., Tutuncuoglu, B., Yan, K., Brown, H., Zhang, Y., Tan, D., Gamalinda, M., Yuan, Y., Li, Z., Jakovljevic, J., et al. (2016). Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 534, 133-137.
16. Li, N., Zhai, Y., Zhang, Y., Li, W., Yang, M., Lei, J., Tye, B.K., and Gao, N. (2015). Structure of the eukaryotic MCM complex at 3.8 A. Nature 524, 186-191.
* indicates co-first authors, # indicates co-corresponding authors
